The Ghosh lab studies how virulence factors of bacterial pathogens interact with their mammalian host cell targets to cause infectious diseases. The lab uses structural biology (i.e., X-ray crystallography, cryo-electron microscopy, and NMR) and biochemistry to generate mechanistic hypotheses, and reverse genetics combined with mammalian cell or animal models of disease to test such hypotheses. The lab is currently focused on mechanisms used by Streptococcus pyogenes M and M-like protein to evade opsonophagocytic killing by the immune system, and by diversity-generating retroelements (DGRs) to create unparalleled levels of protein sequence variation in microbial genomes.
Research
Surface proteins of Strep A
Group A Streptococcus (S. pyogenes or Strep A) remains a major public health threat. This widespread gram-positive bacterial pathogen causes acute invasive diseases and gives rise to severe autoimmune sequelae, and is responsible for morbidity and mortality on a global scale. An essential virulence factor of Strep A, the antigenically variable M protein, enables the bacterium to evade opsonophagocytic killing by the immune system. The M protein confers this indispensable function of phagocyte resistance by recruiting specific soluble human proteins to the GAS surface that block the deposition of the major opsonin C3b as well as antigen-specific opsonic antibodies. In some Strep A strains, an M-like protein serves this function. We are investigating the structural biology and biochemistry of interactions of M and M-like proteins with human targets, and testing functional hypotheses generated by our structural studies through microbiological and immunological experiments. X-ray crystallography has been the prime methodology applied to structural investigations, but others (e.g., EM, NMR, CD) have been applied to address specific questions. The ultimate goal of our studies, which are carried out in collaboration with Victor Nizet, is to provide essential guidance to designing anti-virulence and vaccination strategies against Strep A.
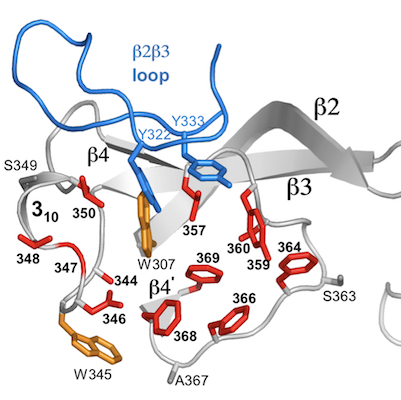
Massive sequence variation in retroelement-encoded proteins
Diversity-generating retroelements (DGRs) are unique and unparalleled generators of massive protein sequence diversity. These elements are prevalent in the microbial ‘dark matter’, which appear to comprise a major fraction of microbial life, and are widespread in the human virome and microbiome. The only other example in the natural world of massive protein sequence variation occurs in the vertebrate adaptive immune system, in which variation enables the recognition of novel targets and consequent adaptation to dynamic environments. A similar benefit appears to be provided by DGRs. DGRs diversify proteins through a fundamentally different mechanism than the vertebrate immune system, and reaches a scope (up to 1030 possible sequences) greatly exceeding that of the vertebrate immune system. In DGRs, diversification arises from genetic information being transmitted unfaithfully for one specific base, adenine, and faithfully for the others. This occurs during reverse transcription of genetic information from RNA to cDNA, and the specificity to adenine shapes the pattern of protein functional variation. This selective infidelity to adenines is the central hallmark feature of DGRs. Selectivity infidelity is unique in biology and we are investigating how it occurs.